Welcome to the FDS Database
Homologous genes (Orthologs) originated from the same ancestor and were separated when the ancestor diverging into two species. These genes are usually regard as playing the same role in different species, which is the key of experiments successfully carried out in animal models before applying to human. However, with the natural selection and evolution, the function of homologous genes might vary with species. Herein we present an index for measuring the functional divergence of homologous gene, FDS, which performed functional enrichment analysis for microRNAs regulating the human-mouse homologous gene and calculated cosine distance for the enrichment significance of function terms. We presented an algorithm and developed an online tool, FDS, for users to query the gene FDS and obtain the same and different function of homologous gene in detail.
Currently, users can
- query the functional divergence score (FDS) value of homologous gene;
- query the same and different function of homologous gene for human and mouse;
- download all data in the database;
Statistics
The current version of the FDS database integrates 15505 human/mouse homologous genes. For one gene, the regulatory element - miRNAs are predicted with 3 tools, including TargetScan, miRanda, and PITA.
Contact Us
Dr Qinghua Cui, 38 Xueyuan Rd, Department of Biomedical Informatics, Peking University Health Science Center, Beijing 100191, China
Email: cuiqinghua@hsc.pku.edu.cn
Citing FDS
Cui C, Zhou Y and Cui Q. Defining the functional divergence of orthologous genes between human and mouse in the context of miRNA regulation. Brief Bioinform (accepted). 2021.
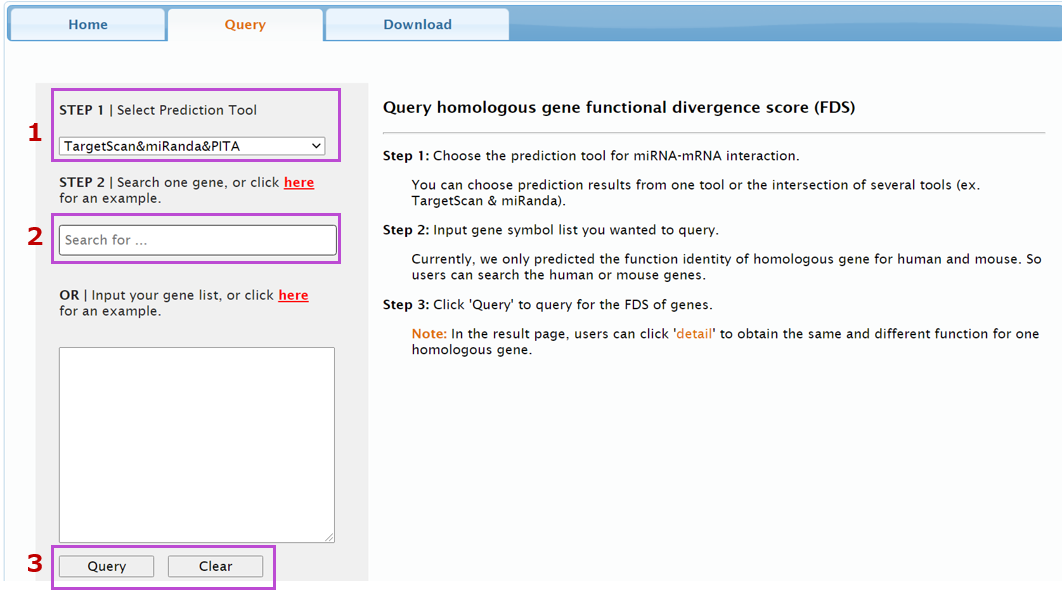
Query homologous gene functional divergence score (FDS)
Step 1: Choose the prediction tool for miRNA-mRNA interaction.
Step 2: Input gene symbol list you wanted to query.
Step 3: Click 'Query' to query for the FDS of genes.
The data are available as shown below:
The FDS values for homologous genes from all prediction tools are available here. (2MB)
The predicting miRNAs for regulating homologous genes with TargetScan are available here. (125MB)
The predicting miRNAs for regulating homologous genes with miRanda are available here. (50MB)
The predicting miRNAs for regulating homologous genes with PITA are available here. (125MB)
The predicting miRNAs for regulating homologous genes with intersection of three tools are available here. (47MB)
The FDS website could be used to query the FDS value and divergent functions and diseases for human and mouse orthologous gene. Hence, users only need to prepare an interested gene symbol or multiple gene symbols to access to the results.
Query: One gene symbol
1. Choose the prediction tool for miRNA-mRNA interaction.
You can choose prediction results from one tool (including TargetScan, miRanda and PITA) or the intersection of several tools (ex. TargetScan & miRanda, TargetScan & miRanda & PITA).
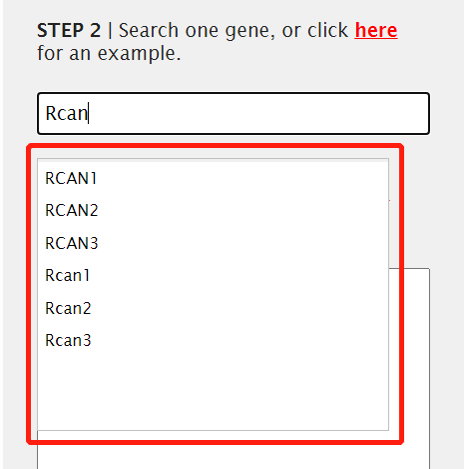
2. Input one gene symbol you wanted to query.
We used the fuzzy-match model to search the gene. When users input several front letters in the textbox, the website can match the eligible genes and pop a dropmenu to exhibit them. Currently, we only predicted the functional divergence of homologous gene for human and mouse. So users can search the human or mouse genes.
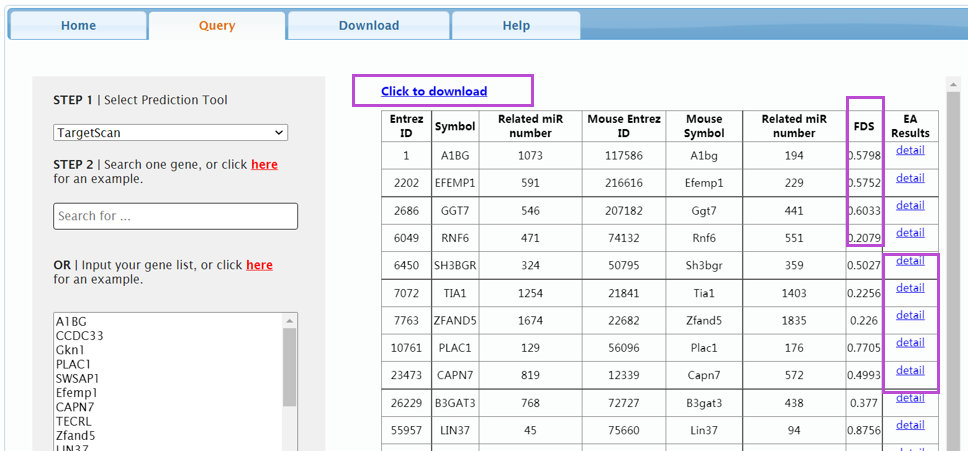
3. Click 'Query' to query for the FDS value of a gene.
Query: Multiple gene symbols
If users want to query the FDS values of multiple genes, they could input many gene symbols separated by line breaks. Similar to the single gene symbol, users must firstly select a prediction tool for miRNA-mRNA interaction. Next, in the textarea, users can input the prepared symbols.
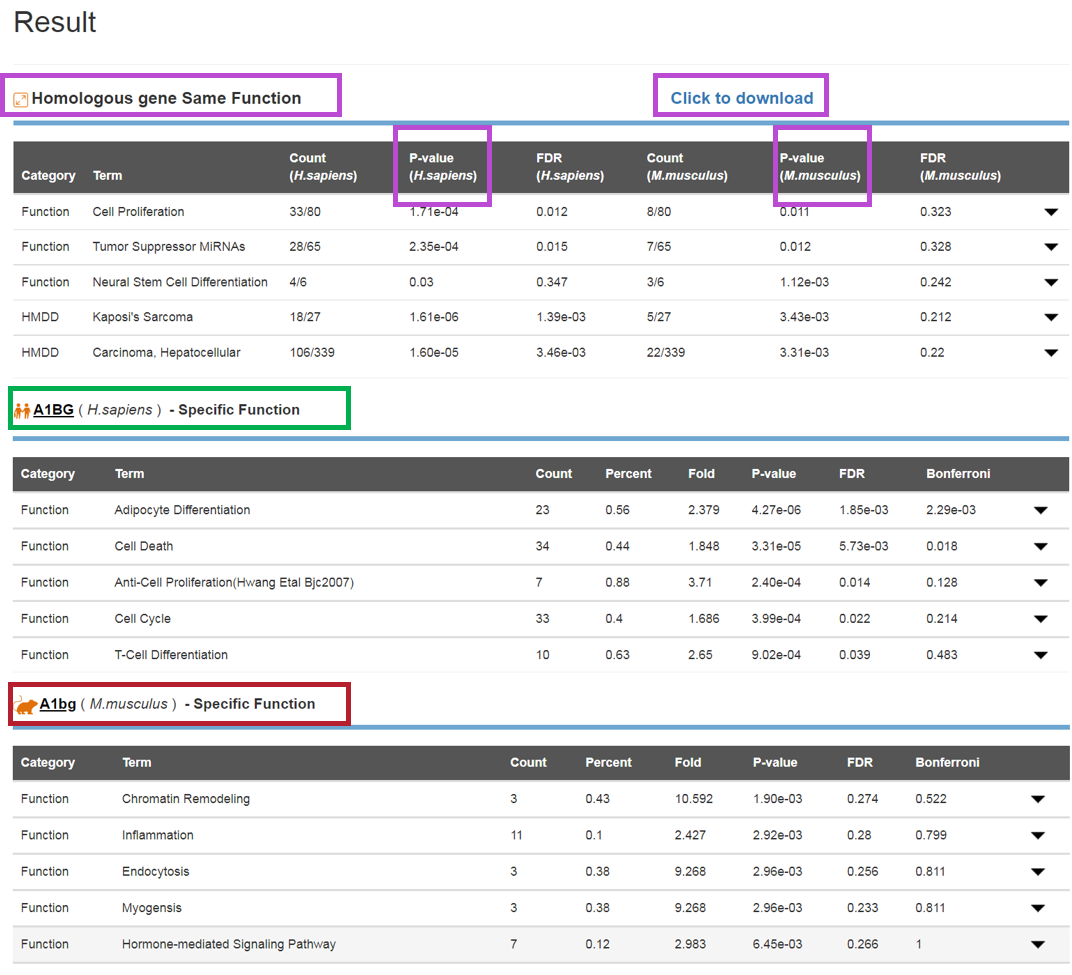
The Query Result Page
The result will be shown in Query page. The human and mouse orthologous genes’ Entrez ID, symbol, the number of regulating miRNAs and FDS value are listed in the result page. Users can browse the accordance and divergent functions and diseases of the orthologous gene by clicking the 'Detail' link in the corresponding entry.
The Detail Page
The detailed result will be shown in a new page and contains the same and different functions and diseases for an orthologous gene. The result will be partitioned into three parts, including the same functions and diseases of the human and mouse orthologous gene pair, the specific functions and diseases of human gene and the specific functions and diseases of mouse orthologous gene. In the first part, the result listed the significant function and disease terms, the number of miRNAs, regulating the human orthologous gene, involving the corresponding term, the p-value of hypergeometric distribution, which is always used for the functional enrichment analysis, and FDR. In the specific part, we listed the term, miRNA number, miRNA percent, fold, p-value, FDR and Bonferroni-adjusted p-value, similar to TAM. We set the threshold of p-value ≤ 0.05 as the significantly correlated functions and diseases of the orthologous gene.