LncTar: an efficient tool for predicting RNA targets of lncRNAs
Long noncoding RNAs (lncRNAs) play important roles in regulation of many biological processes. LncRNAs have a diversity of functions through various mechanisms by binding diverse molecules, such as proteins, RNAs, and DNA. Among their binding molecules, RNA represents one of the most popular targets. Therefore, given the big number of lncRNAs, it becomes urgent and important to predict lncRNA-RNA interactions by developing bioinformatics methods. The sequences of lncRNAs are usually long and not fully complementary to the target sequences. Their regulatory functions are functioned by establishing stable joint structures with target sequences. This make is difficult for predicting lncRNA-RNA interactions. For this purpose, here we presented an efficient tool, LncTar.
LncTar is a software for predicting lncRNA-RNA interactions by means of free energy minimization. LncTar utilized a variation on the standard "sliding" algorithm approach to calculate the normalized binding free energy (ndG) and found the minimum free energy joint structure. The ndG was regard as a cutoff which determining the paired RNAs as either interacting or not. Of course, LncTar is not specific for lncRNAs but also can be used for predicting putative interactions among various types of RNA molecules, such as mRNA, noncoding RNAs including lncRNAs, pre-miRNAs, and other types of noncoding RNAs.
History
November-10, 2015, LncTar web server was released.
September-01, 2015, the calculation of ndG was improved.
September-01, 2014, LncTar standalone software was released.
Related articles
Li J, Ma W, Zeng P, Wang J, Geng B, Yang J, and Cui Q. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Briefings in Bioinformatics 16(5) 2015.
Copyrights
Jianwei Li and Qinghua Cui. The software and source codes can only be used for academic purpose. For commercial usage, please contact with Dr.Qinghua Cui.
Contact us
Dr. Qinghua Cui, 38 Xueyuan Rd, Department of Biomedical Informatics, Peking University Health Science Center, Beijing 100191, China
Email: cuiqinghua@hsc.pku.edu.cn
Homepage: http://www.cuilab.cn/
LncTar is a software for predicting lncRNA-RNA interactions by means of free energy minimization. LncTar utilized a variation on the standard “sliding” algorithm approach to calculate the normalized binding free energy (ndG) and found theminimum free energy joint structure. The ndG was regard as a cutoff which determining the paired RNAs as either interacting or not. Of course, LncTar is not specific for lncRNAs but also can be used for predicting putative interactions among various types of RNA molecules, such as mRNA, noncoding RNAs including lncRNAs, pre-miRNAs, and other types of noncoding RNAs.
The detailed usage of LncTar is as followings:
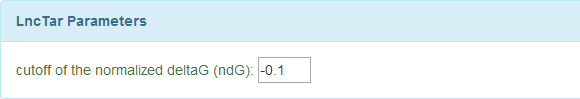
[1] LncRNA-RNA Interaction Parameters Setting
[2] Input
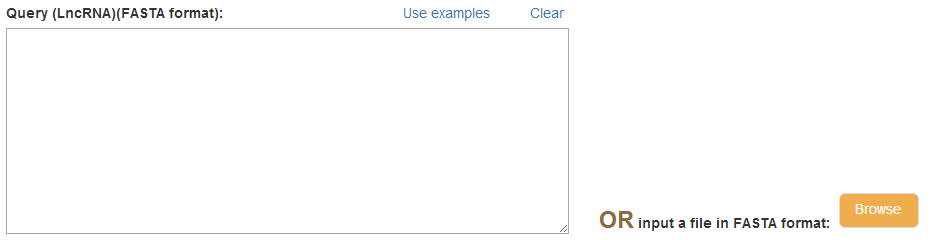
In this section, you are requested to input the sequence of lncRNA(s) in the format of FASTA, and you have two ways to input the sequence: paste a sequence or input a file, you only need to enter one of them.
Attention1: Make sure that the sequence doesn't contain other characters except "A","T","C","G","U".
Attention2: LncTar allows to input several sequences, but for efficiency, we suggest you test one by one.
Attention3: Suggest you click "Use examples" to see the standard format.
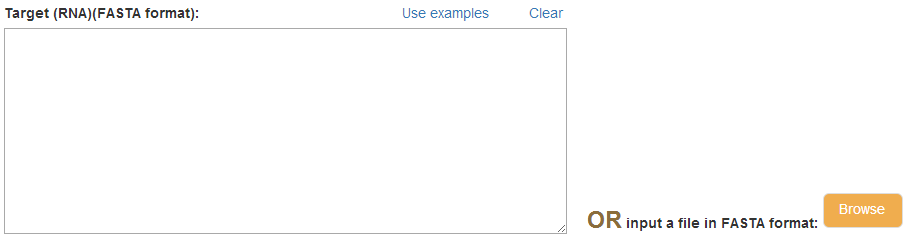
In this section, you need to input the sequence of RNA(s) that you want to test whether it may react with the lncRNA(s) you have entered.
[3] Run
click the "Submit" button and wait patiently, because it will take several minutes!