

News:
The tool for gene pattern discovery by enrichment analysis of their microRNA regulators ia a web-accessible program, which is used to mine potential patterns for one single gene, multiple genes, and KEGG pathway genes based on miRNA set enrichment analysis of the miRNAs regulating given genes. For any given gene, multiple genes, or one pathway, miR2Gene is able to:
Contact Us
Dr Qinghua Cui, 38 Xueyuan Rd, Department of Biomedical Informatics, Peking University Health Science Center, Beijing 100191, China
Email: cuiqinghua@hsc.pku.edu.cn
Homepage: http://www.cuilab.cn/
Best viewed with Mozilla Firefox 3.0+ or Internet Explorer 7.0+, and note that SVGview plugin must be installed if you use IE to view the pathway figures.
Step 1: put your gene name through left panel.
An example:
| ABL2 | OFFICIAL_GENE_SYMBLE |
| TP53 | OFFICIAL_GENE_SYMBLE |
| DQ305855 | GENBANK_ACCESSION |
| NM_002268 | REFSEQ_MRNA |
| ENSG00000168779 | ENSEMBL_GENE_ID |
| ENST00000264382 | ENSEMBL_TRANSCRIPT_ID |
| 6478 | ENTREZ_GENE_ID |
| uc003yuk.2 | UCSC_GENE_ID |
Step 2: select the right gene symble type(as step 1 example shows).
Step 3: choose a microRNAs target predict method
Step 4: submit the request.
Step 1: put your gene name through left panel.
An example:
| ABL2 | OFFICIAL_GENE_SYMBLE |
| TP53 | OFFICIAL_GENE_SYMBLE |
| DQ305855 | GENBANK_ACCESSION |
| NM_002268 | REFSEQ_MRNA |
| ENSG00000168779 | ENSEMBL_GENE_ID |
| ENST00000264382 | ENSEMBL_TRANSCRIPT_ID |
| 6478 | ENTREZ_GENE_ID |
| uc003yuk.2 | UCSC_GENE_ID |
Step 2: select the right gene symble type(as step 1 example shows)
Step 3: choose a microRNAs target predict method
Step 4: set the threshold value. If you set 2 as the default threshold value, only the mirs which regulate no less than two genes simultaneously will be processed.
Step 5: submit the request
Select one KEGG human pathway to annotate the regulatory genes it contains with the miRNAs sets enrichment analysis results of the interest miRNAs regulate them.
A sample of KEGG pathway analysis result
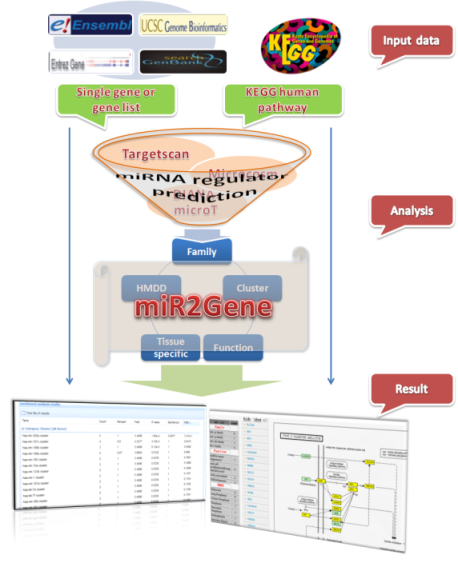
miR2Gene is a web server to explore hidden patterns for protein-coding genes through enrichment analysis of their microRNA (miRNA) regulators. miR2Gene first integrated miRNAs into groups according to prior rules, such as function, associated disease, tissue specificity, family, and cluster. miR2Gene then predicted the miRNAs that regulate given protein-coding genes using several algorithms. Next, an enrichment analysis of miRNA regulators in various miRNA sets is performed. Finally, the enriched miRNA sets are identified for given protein-coding genes. miR2Gene can be used for a single gene, multiple genes, or KEGG pathways.
[1]. Log on miR2Gene web-server
Users can log on the miR2Gene web-server at http://www.cuilab.cn/mir2gene, as shown in Figure 1.
Figure 1. The homepage of miR2Gene
[2]. Pattern discovery for protein-coding genes or KEGG pathways
Here, taking a single gene "ABL2" and KEGG pathway "wnt signaling pathway" as examples.
(1) Analysis of single gene. The analysis procedure is as follows:
Figure 2. The analysis procedure of miR2Gene for single gene
(2) Interpretation of the analysis result for single gene.
Figure 3. The Result (part result) of analysis result for gene ABL2.
The results (miRNA sets) are organized into five categories, miRNA cluster, miRNA family, Function, HMDD (miRNA associated disease), and TissueSpecific (Figure 3). The users can rank the results by values of Count (number of matched miRNAs), Percent (percentage of matched miRNAs in corresponding miRNA set), Fold (The real matched number/expected matched number), P-value, Bonferroni (corrected p value by Bonferroni), FDR (corrected p value by FDR). The significantly enriched miRNA sets are putatively associated with the given protein-coding gene. One important point the users should take care is that because the relationship between genes and their miRNA regulators is negative regulation, the discovered function should be reversed in some times. For example, the most significant miRNA set in Functional category is "miRNA tumor suppressors", which means the gene "ABL2" have very strong tendency of oncogene. Indeed, ABL2 is an oncogene.
(3) Analysis of KEGG pathways. The analysis procedure is as follows:
Figure 4. The analysis procedure of miR2Gene for KEGG pathway
(4) Interpretation of the analysis result for KEGG pathway.
Figure 5. The Result (part result) of analysis result for Wnt signaling pathway.
A new page will be shown for the result of KEGG pathway analysis (Figure 5). The enriched miRNA sets (FDR<=0.05) are listed and organized by the above five categories (red "1" in Figure 5). The number of genes having corresponding enriched miRNA sets in selected pathway is also listed (red "2" in Figure 5). Clicking corresponding miRNA set, the significant genes will be highlighted in the pathway. A function to hide or show all highlighted genes in the pathway is also provided (red "3" in Figure 5). Furthermore, all genes in the pathway are also listed (red "4" in Figure 5). Clicking any of the listed genes will show the significant miRNA sets. After finishing tuning all the parameters, the resulted figure of the pathway will be shown (red "5" in Figure 5), in which, the significant genes are highlighted in yellow color.
[3]. Reference & Contact Information
Qiu et al. miR2Gene: Gene pattern discovery by enrichment analysis of their microRNA regulators. (submitted).
Dr. Qinghua Cui, http://www.cuilab.cn/, cuiqinghua@hsc.pku.edu.cn
Citation: Chengxiang Qiu et al. miR2Gene: Pattern discovery of single gene, multiple genes, and pathways by enrichment analysis of their microRNA regulators.
BMC Systems Biology (in press).