A tool for annotations of weighted human microRNAs
Welcome to the wTAM
With the development of high-throughput sequencing, numerous miRNA-omics data have been accumulated. It is important to annotate the miRNA-seq data with high quality information. There exist some tools for annotating miRNA data, such as, TAM, miEAA. These above methods regard that all miRNAs have the same importance. However, the miRNAs exhibit differentiate weight in various biological processes. Given to the weighted score of miRNAs, we introduced wTAM, a tool for annotation of weighted miRNAs.
Statistics:
There are 8 well-annotated miRNA sets for further analysis, including miRNA cluster, miRNA family, Function, HMDD, Transcript Factor (TF), tissue specificity, cell specificity and Environment Factor (EF). In addition, we collected 17 miRNA importance scores, covering a series of miRNA features, for example, miRNA conservation, miRNA expression.
History:
August, 2021, wTAM web server was released;
July, 2021, wTAM web server was tested;
June, 2021, the accuracy of wTAM algorithm was tested;
April, 2021, wTAM algorithm was built;
The miRNA sets and miRNA important score files are available here!
User Guides
We provided instructions for the main functions in webserver. Users could click the part of their interest in the following navigator.
Analysis (weighted enrichment analysis)
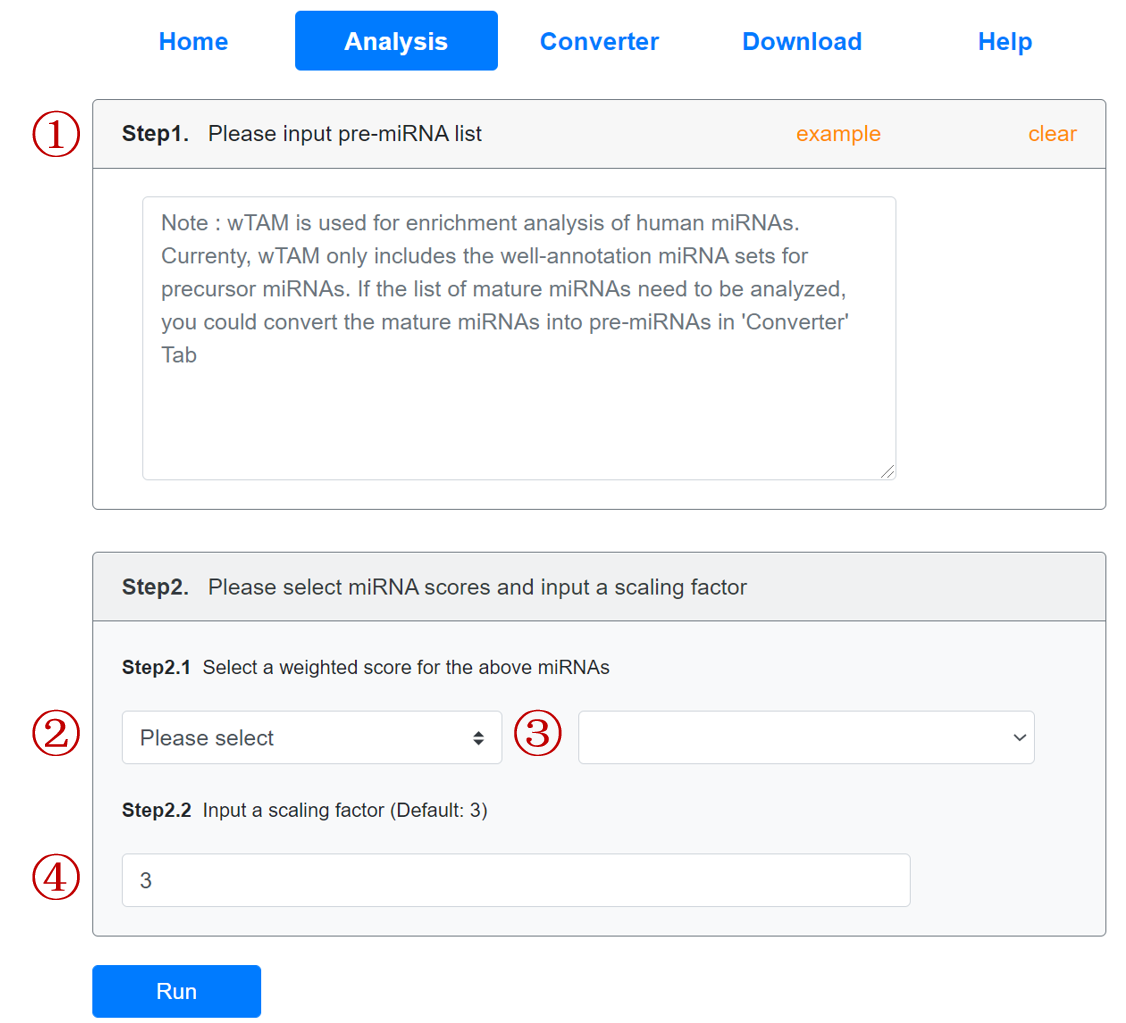
Input the list of pre-miRNA names that you want to analyse:
- Input the pre-miRNAs. The user can click the 'example' button for an example.
-
Select weighted scores for miRNAs.
-
Firstly, users need to select the class in the left dropdown menu of 'Step2.1'; Subsequently, click the right dropdown menu to select the detailed scores. When users select the 'User defined', they should input the pre-defined weighted scores for all miRNAs in csv format on the right textarea of 'Step2.1' panel.
How to select the weighting score?
We provided 5 classes of weighting scores and separately introduce the application context. If the given miRNA lists are from disease-related circumstances, we recommend the miRNA importance score, which is based on the association of miRNA and diseases. The expression score is used for exploring the pathways or biological processes under a specific tissue. The conservation score is suggested to dissecting the more conserved pathways. The downstream score characterized by the number of mRNA targeted by miRNAs, is used to explore impacts on downstream pathways. Similarly, the upstream score represented by the number of transcription factors regulating miRNAs could be used to screen the functions influenced by the upstream exchanges of miRNA. Additionally, we also support users to upload self-defined weightings with biological meaning for their interested miRNA lists.
- Users need to input the scaling factor of factor number. The default scale is 3.
-
Analysis Result
The result will be shown in a new page. The parameters users selected are shown in the result page. The miRNA set category, term, count, percent, P-value of wTAM, FDR, P-value of TAM, and the significant term founded by wTAM with star label will be shown in the web page.
Meanwhile, users could click the “+” signal for the detail of term. In the detail area, the hit miRNAs and missed miRNAs are shown.
We have supported visulization for the result. Users could set the threshold of signigicantly enriched terms and set the top N terms in the Bubble plot or Bar chart.
In addition, full screen, select the result columns, export the result and search keywords in the result table are available in the toolbar in the top right corner.


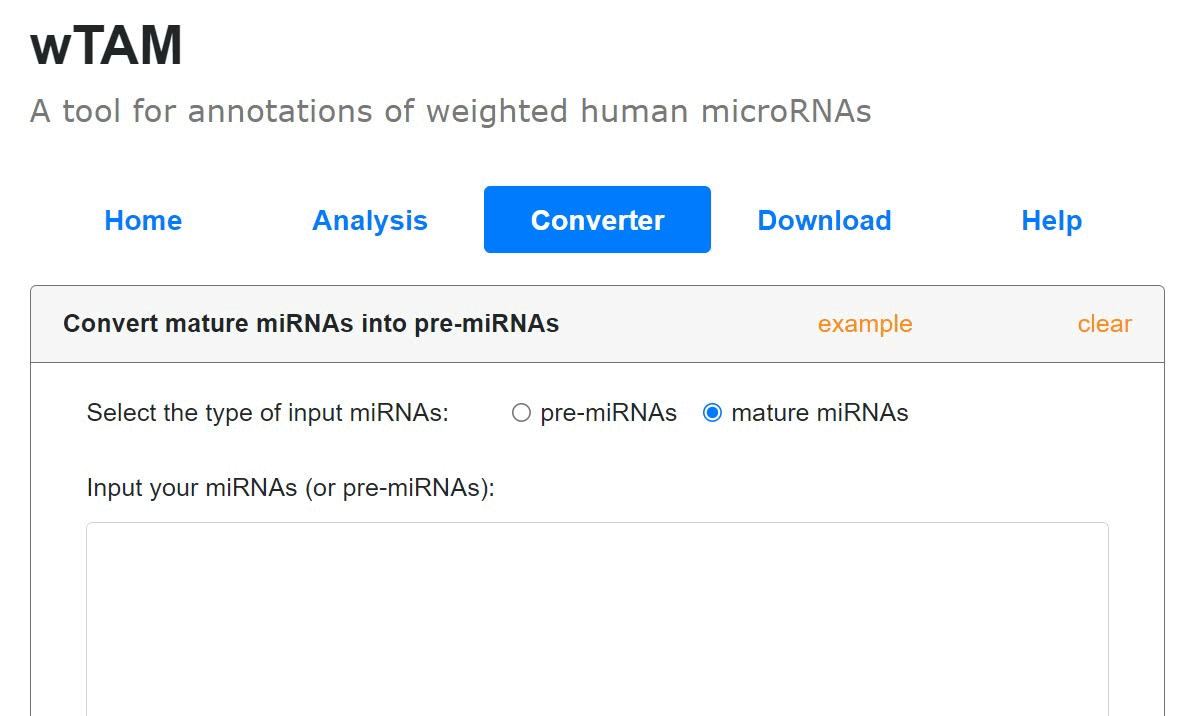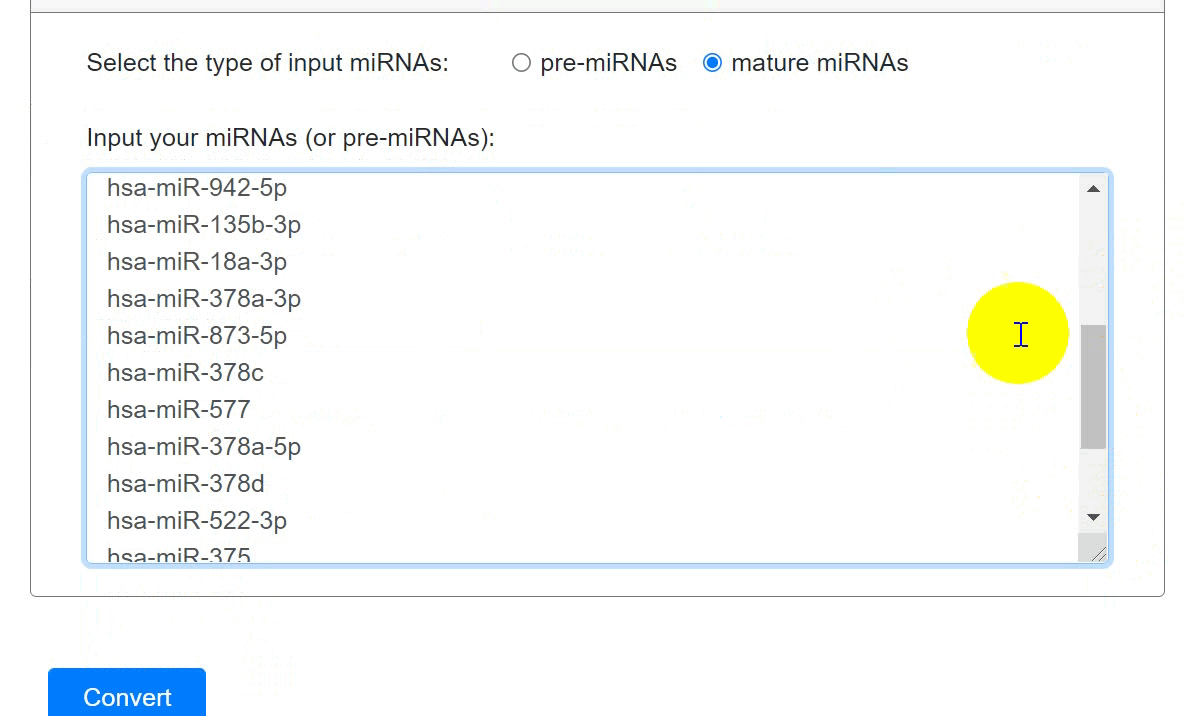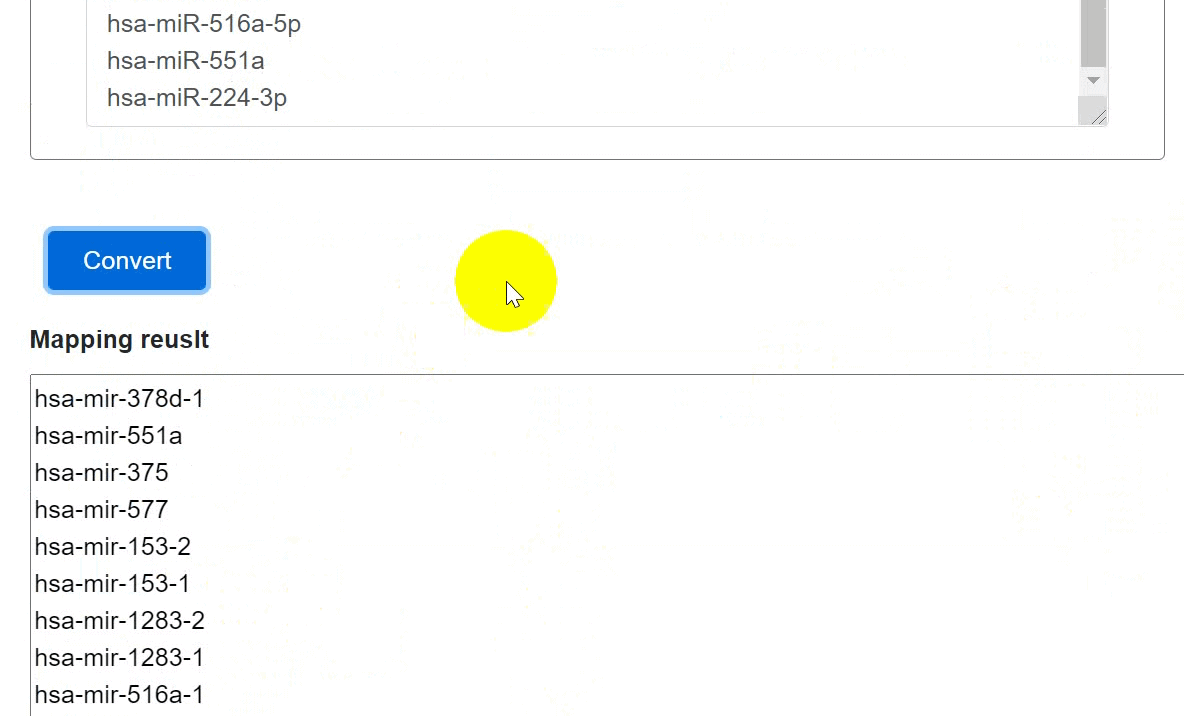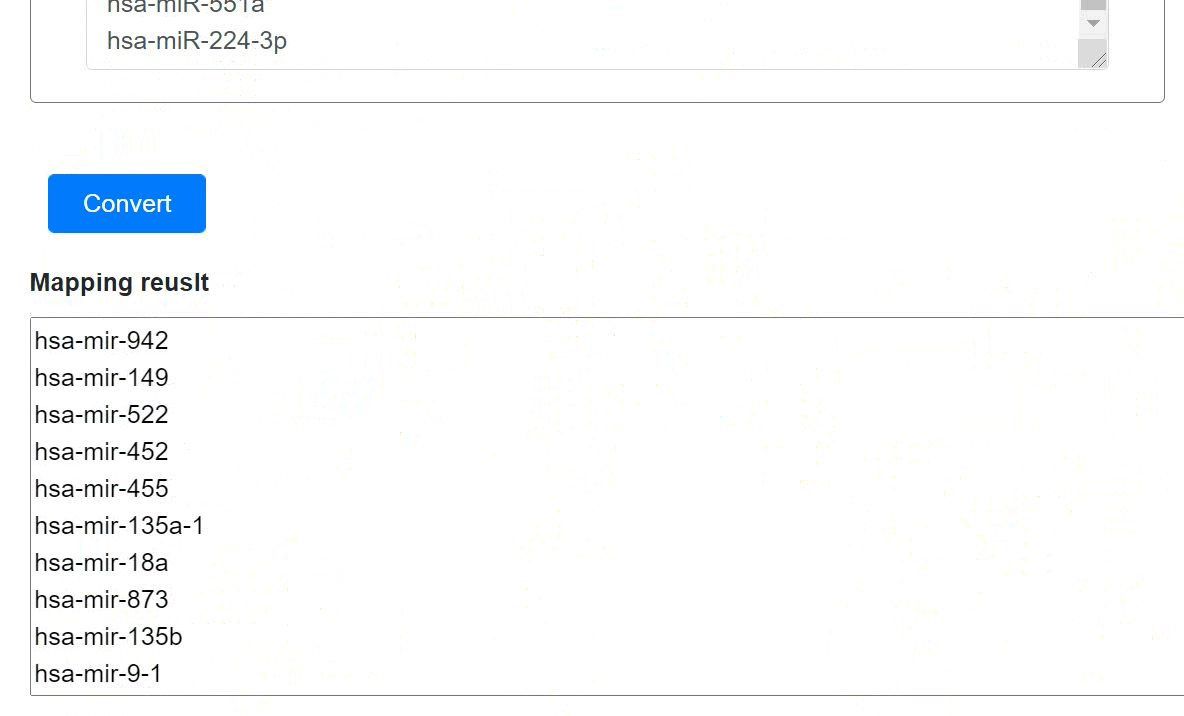
Converter
Users could convert precursor miRNAs into mature miRNAs and vice versa:
- Choose the type of input miRNAs.
- Input miRNAs names.
- Click the 'Convert' to submit the miRNAs.