miES: a tool for microRNA Essentiality Score
Welcome to miES
MicroRNAs (miRNAs) are one class of small noncoding RNA molecules, which regulate gene expression at the post-transcriptional level and play important roles in health and disease.
To dissect the critical miRNAs in miRNAome, it is needed to predict the essentiality of miRNAs,
however, bioinformatics methods for this purpose are limited.
Here we propose miES, a novel algorithm, for the prioritization of miRNA essentiality.
miES (microRNA Essentiality Score) implements a novel machine learning strategy based on learning from positive and unlabeled samples.
miES uses sequence features of known essential miRNAs and performs miRNAome-wide searching for new essential miRNAs.
Contact us
Dr. Qinghua Cui
38 Xueyuan Rd, Department of Biomedical Informatics,
Peking University Health Science Center,
Beijing 100191, China
Email: cuiqinghua@hsc.pku.edu.cn
Dr. Lin Gao
School of Computer Science and Technology,
Xidian University,
No2 Taibai South Road, Xi'an, Shaanxi Province, 710071, China
Email: gao@mail.xidian.edu.cn
miES source code in R is available here
Introduction
MicroRNAs are one class of small non-coding RNAs, which has attracted more and more attention in recent years. Given the big number of miRNAs, it becomes important to quantify the importance of each miRNA. Therefore we proposed a logistic regression model, miES (microRNA essentiality score), to quantify the essentiality of miRNAs. miES uses the sequence information of a miRNA based on data of 77 essential mice miRNAs from Bartel’s review paper(Bartel, 2018) and predicted novel data in miRBase and miRCarta databases.
Visit miES
miES is available at http://www.cuilab.cn/miES.
Prepare input data
For a miRNA, miES needs the sequence of its pre-miRNA and its mature miRNA(s). In single prediction,the pre-miRNA sequence should be arranged as FASTA format and inputted into the first input panel. The mature miRNA sequence(s) also should be arranged as FASTA format and inputted into the second input panel. And in multiple prediction, per line should be one miRNA which contains pre-miRNA name, pre-miRNA sequence and mature-miRNA sequence(s) seperated by tab.
Predict
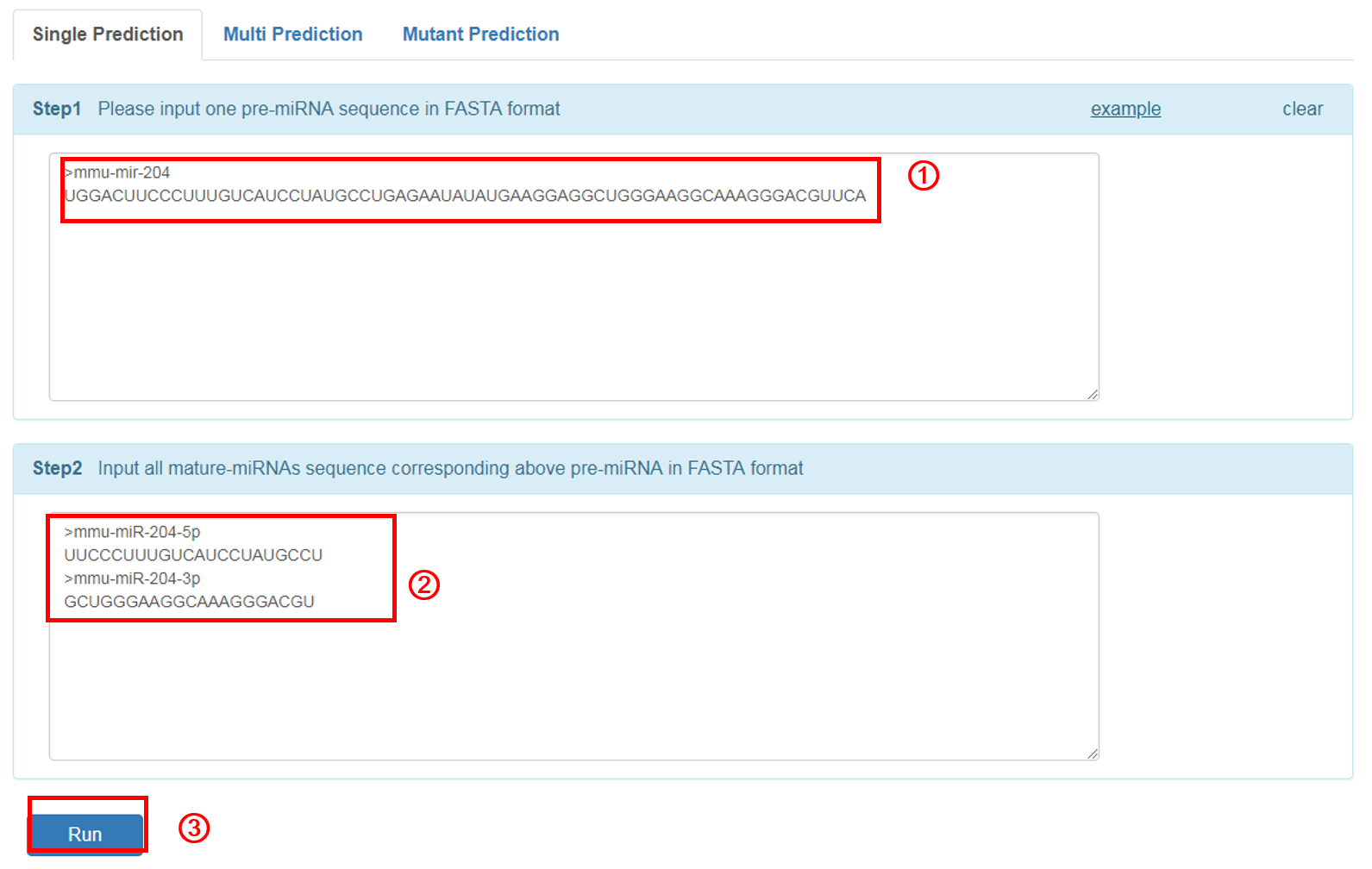
Single Prediction: input miRNA sequence that you want to predict:
1. Input the pre-miRNA sequence in fasta format. The user can click the 'example' button for an example sequence.
2. Input all mature miRNA sequence(s) of the above pre-miRNA in fasta format.
3. Click 'RUN' to submit the sequence.
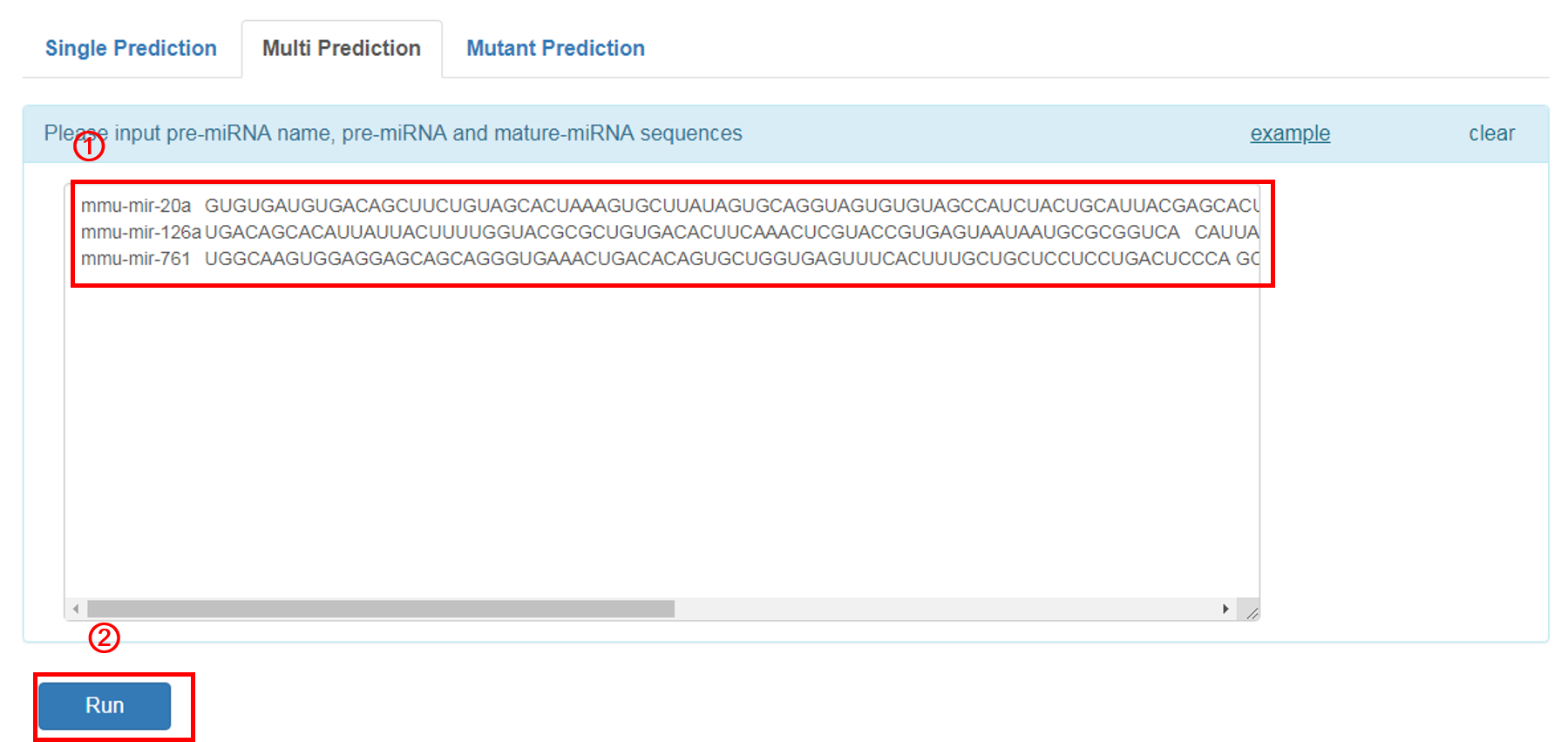
Multiple Prediction:
1. Input the pre-miRNA name, pre-miRNA and corresponding mature-miRNA sequnces in example format. The user can click the 'example' button for an example sequence.
2. Click 'RUN' to submit the sequence.
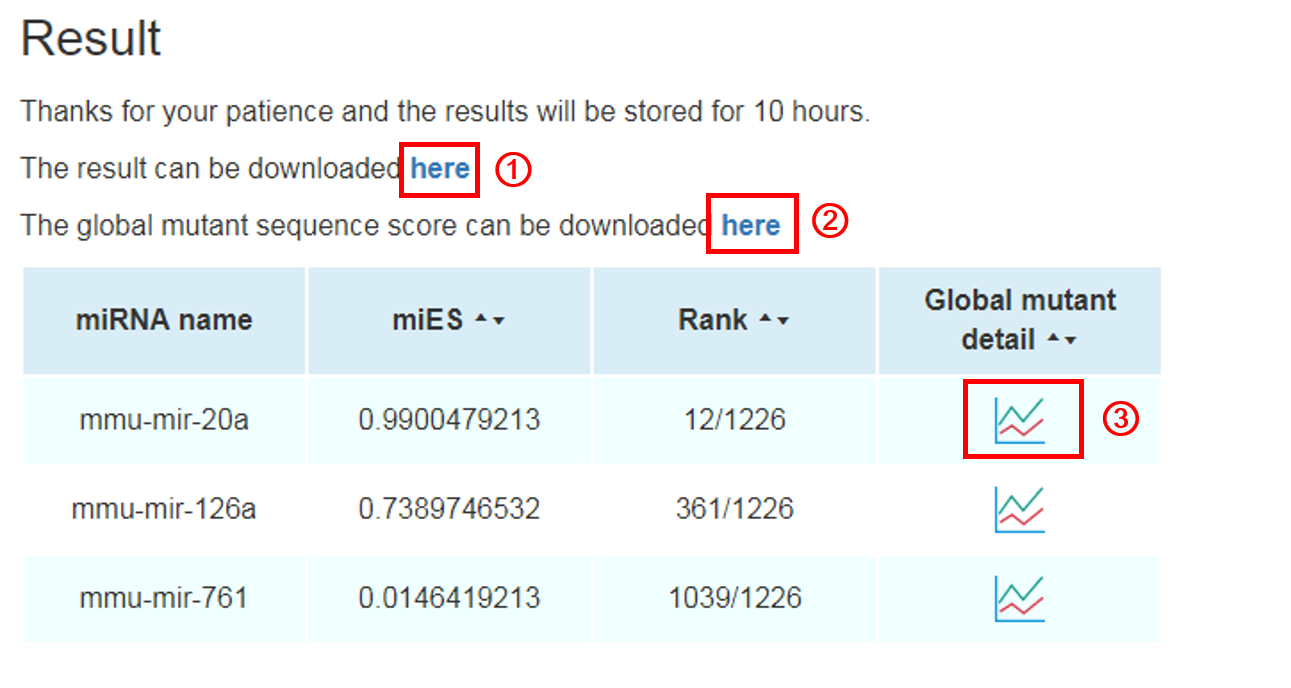
Result:
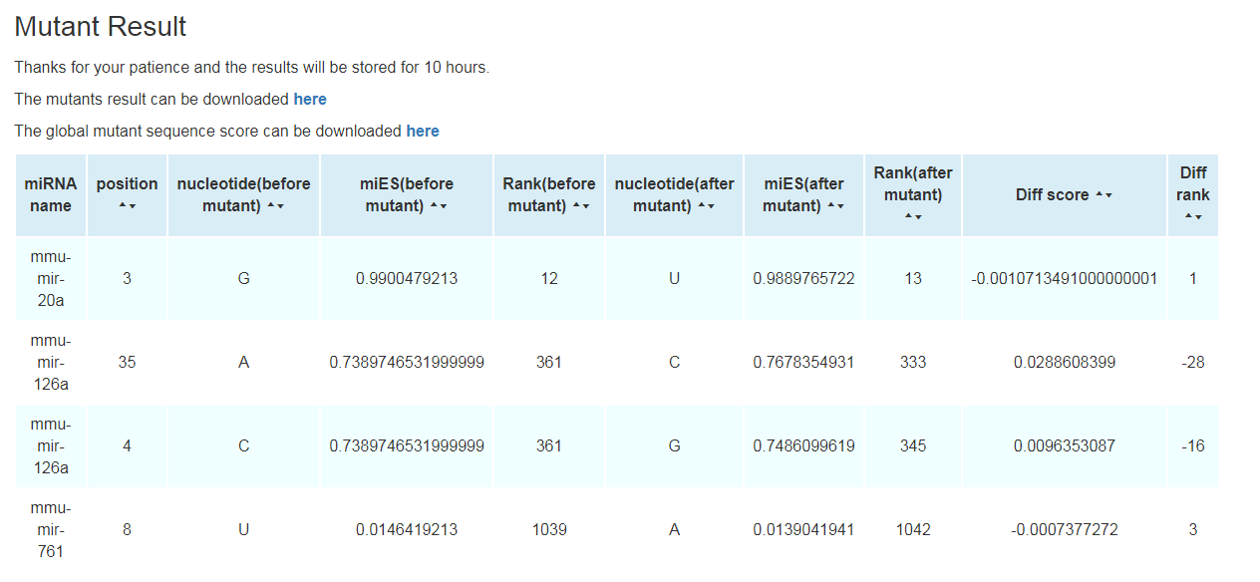
The result will be shown in a new page. The miRNA name, miES, and its relative rank in all miRNAs will be shown in the web page. Meanwhile, two files can be downloaded in the result page. The result file 1 contains miRNA name, miES, and its relative rank. We also putatively mutate each nucleotide in the miRNA sequence into the other three nucleotides and calculate all of the miES and rank for each mutant in each position of nucleotide. These results are available in the result file 2. In addition, the visualization of single nucleotide mutant result can be found by graphical button.

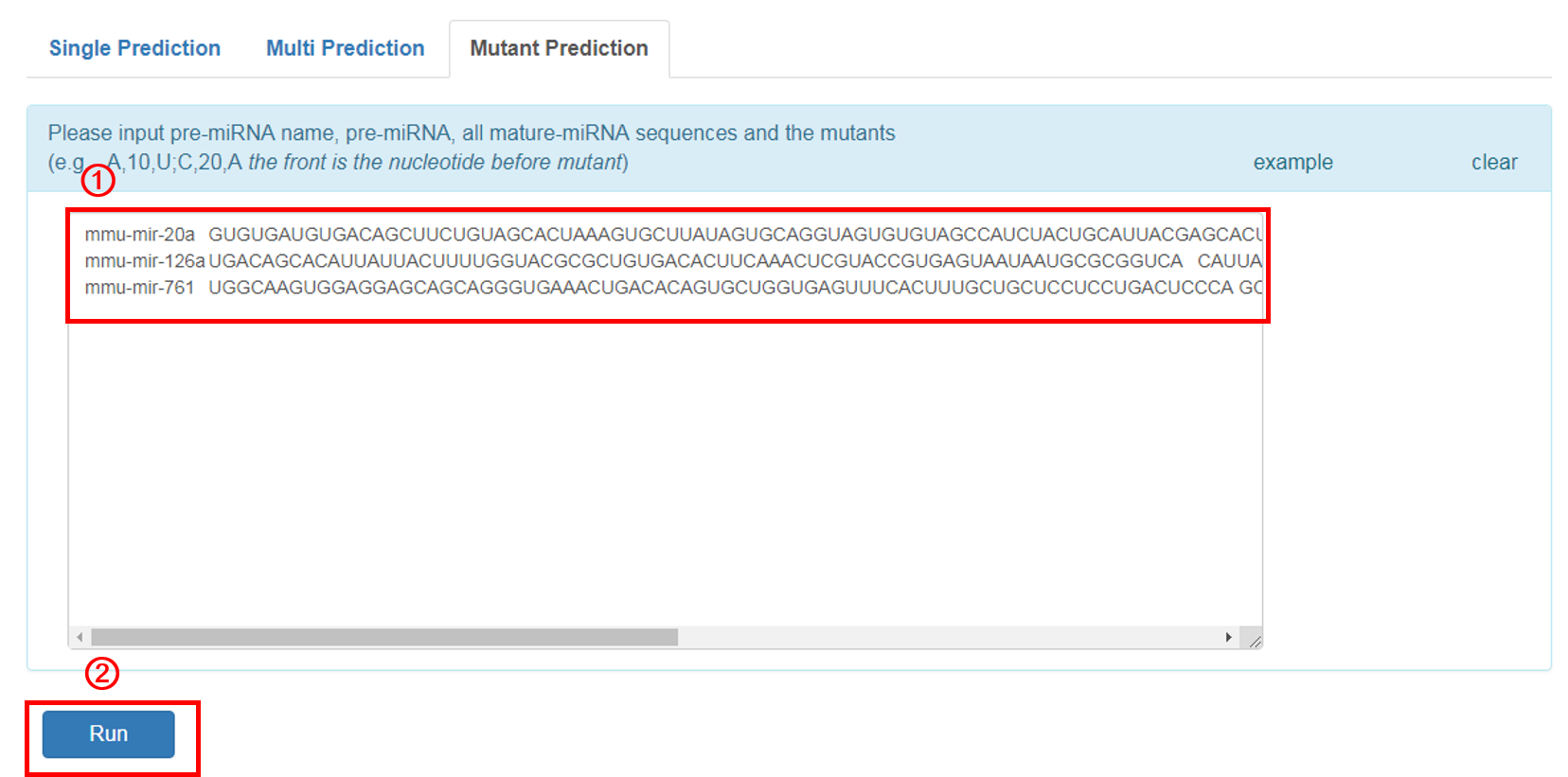
Mutant Prediction:
The user can input some single mutants they care about in some miRNAs.
1. Input the pre-miRNA name, pre-miRNA, corresponding mature-miRNA sequnces and and the mutants in example format. The user can click the 'example' button for an example sequence.
2. Click 'RUN' to submit the sequence.
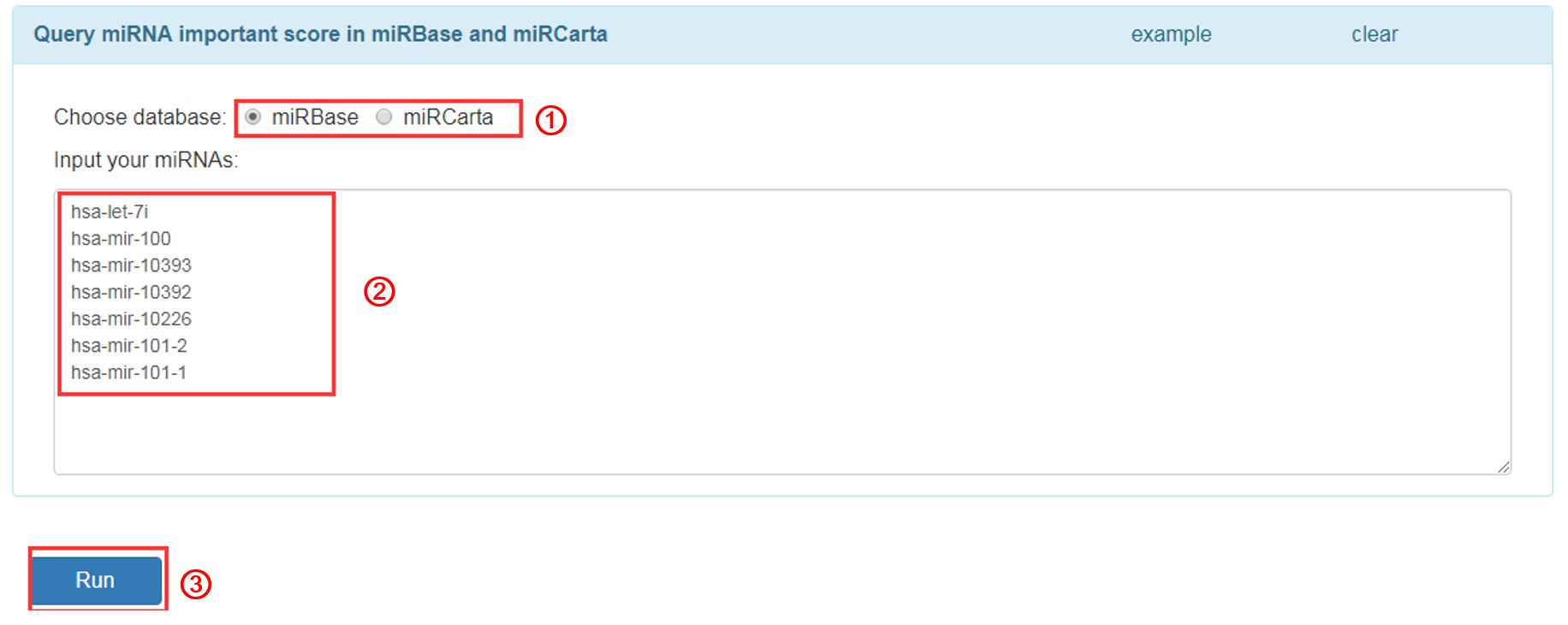
Query
Input miRNA name that you want to query:
1. Choose one miRNA database, currently we provide two databases, miRBase and miRCarta.
2. Input miRNA names (one miRNA per line).
3. Click 'RUN' to submit the miRNAs.