MIC: MicroRNA Importance Calculator
Welcome to MIC
microRNAs (miRNAs) are one class of small noncoding RNA molecules that mainly regulate gene expression negatively at the post-transcriptional level. It has been shown that miRNAs have important molecular functions and play critical roles in various key biological processes. Therefore, miRNAs are potential molecules in disease diagnosis and therapy. Given the big number of miRNA molecules, it thus becomes emergently needed to evaluate the importance of each miRNA.
MIC (MicroRNA Importance Calculator) is a web server for the above purpose. Using the inputted sequences of pre-miRNA and its mature miRNAs, MIC will calculate the importance score of the given miRNA.
History:
April, 2018, MIC web server was released;
March, 2018, MIC web server was tested;
Februry, 2018, the accuracy of MIC algorithm was tested;
January, 2018, MIC algorithm was built;
November, 2017, sequence features were tested;
Contact us
Dr. Qinghua Cui
38 Xueyuan Rd, Department of Biomedical Informatics,
Peking University Health Science Center,
Beijing 100191, China
Email: cuiqinghua@hsc.pku.edu.cn
The miRNA important score files are available here (6 species, including human, rat, mouse, drosophila, zebrafish and elegans)
The raw data of MIC algorithm are available here
Introduction
MicroRNAs are one class of small non-coding RNAs, which has attracted more and more attention in recent years. Given the big number of miRNAs, it becomes important to quantify the importance of each miRNA. Here we proposed MIC, microRNA importance calculator, for this purpose. MIC uses the sequence information of a miRNA to calculate the importance score of the miRNA. MIC constructed regression model based on data in the HMDD (human microRNA disease database) and miRBase databases, and predicted novel data in miRBase and miRCarta databases.
Visit MIC
MIC is available at http://www.cuilab.cn/mic.
Prepare input data
For a miRNA, MIC needs the sequence of its pre-miRNA and its mature miRNA(s). In single prediction,the pre-miRNA sequence should be arranged as FASTA format and inputted into the first input panel. The mature miRNA sequence(s) also should be arranged as FASTA format and inputted into the second input panel. And in multiple prediction, per line should be one miRNA which contains pre-miRNA name, pre-miRNA sequence and mature-miRNA sequence(s) seperated by tab.
Predict
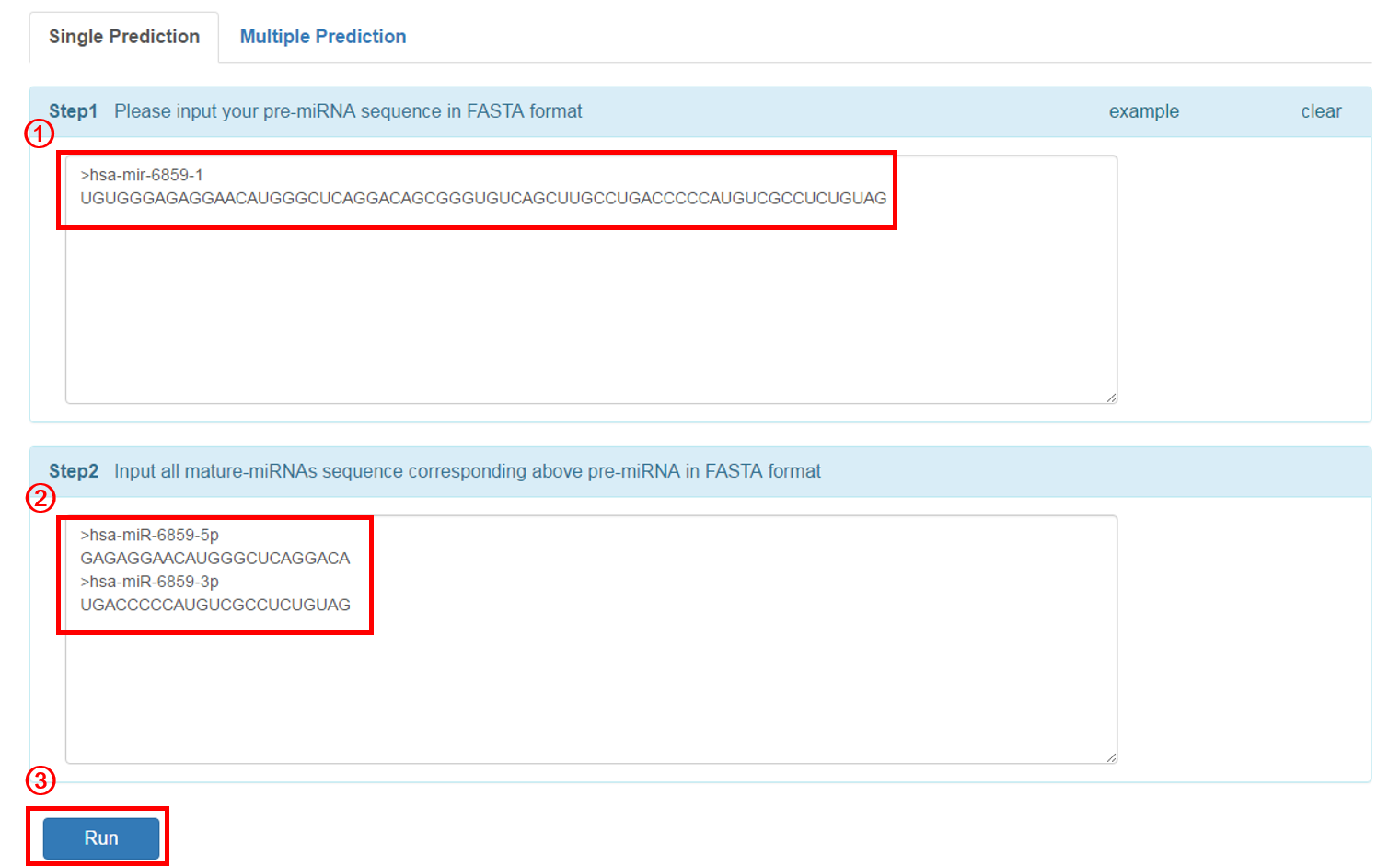
Single Prediction: input miRNA sequence that you want to predict:
1. Input the pre-miRNA sequence in fasta format. The user can click the 'example' button for an example sequence.
2. Input all mature miRNA sequence(s) of the above pre-miRNA in fasta format.
3. Click 'RUN' to submit the sequence.
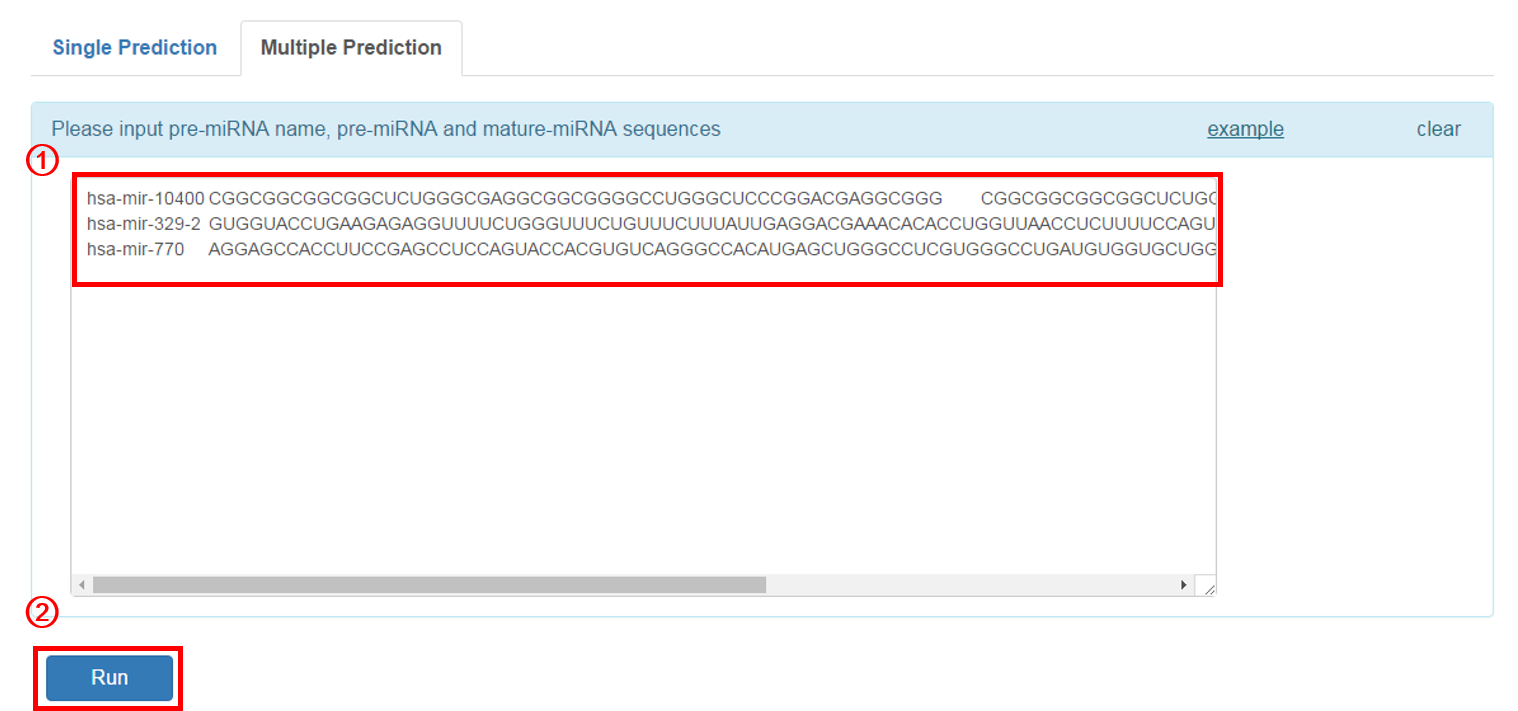
Multiple Prediction:
1. Input the pre-miRNA name, pre-miRNA and corresponding mature-miRNA sequnces in example format. The user can click the 'example' button for an example sequence.
2. Click 'RUN' to submit the sequence.
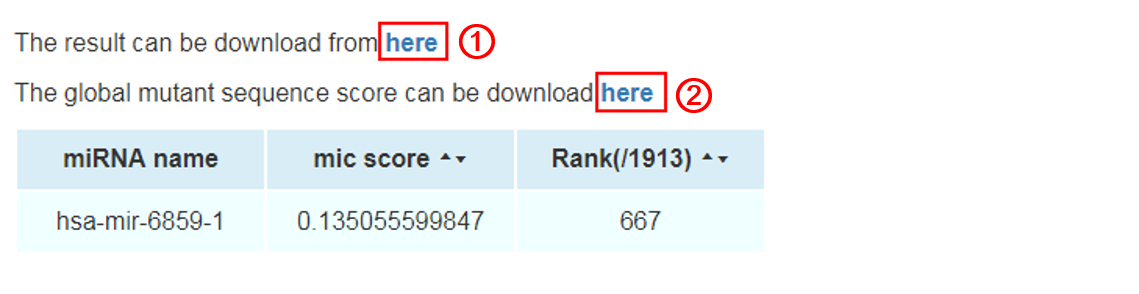
Result:
The result will be shown in a new page. The miRNA name, MIC score, and its relative rank in all miRNAs will be shown in the web page. Meanwhile, two files can be downloaded in the result page. The result file 1 contains miRNA name, MIC score, and its relative rank. We also putatively mutate each nucleotide in the miRNA sequence into the other three nucleotides and calculate all of the MIC score and rank for each mutant in each position of nucleotide. These results are available in the result file 2.
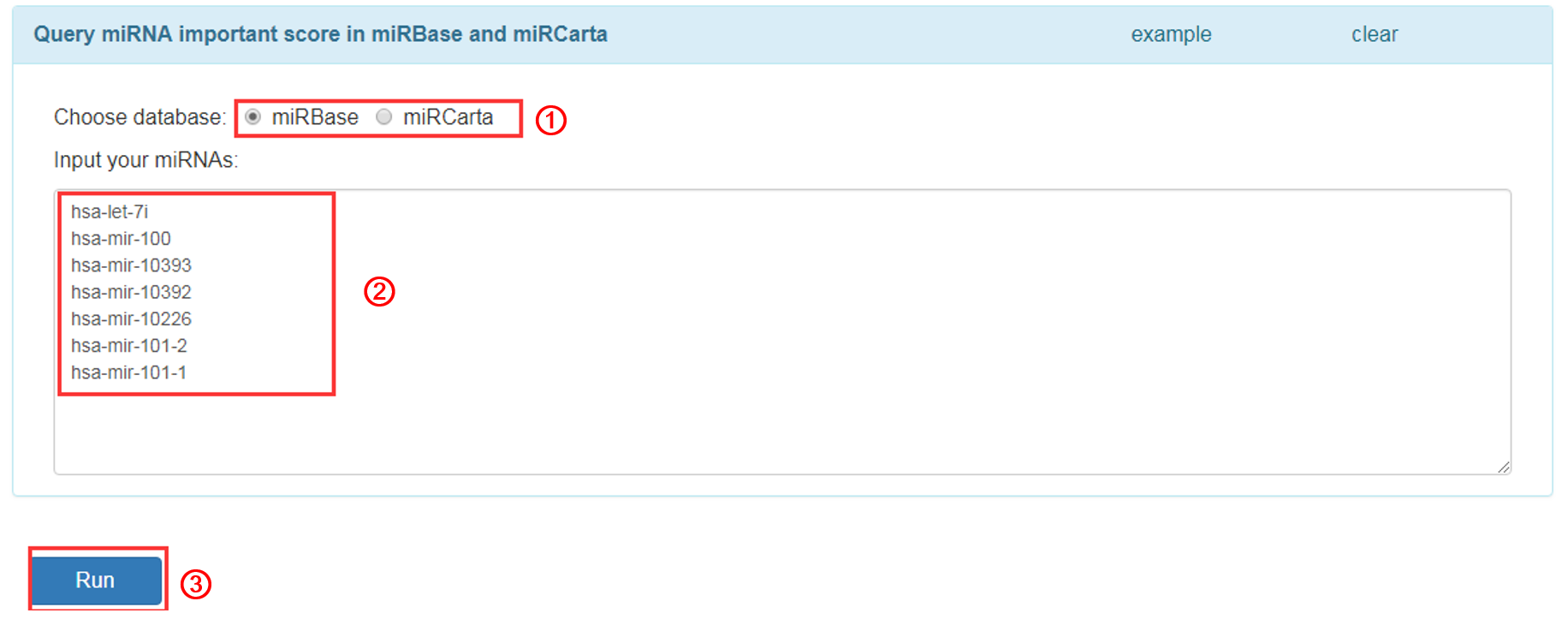
Query
Input miRNA name that you want to query:
1. Choose one miRNA database, currently we provide two databases, miRBase and miRCarta.
2. Input miRNA names (one miRNA per line).
3. Click 'RUN' to submit the miRNAs.