Welcome to PIC!
Introduction
The Challenge
Human essential proteins (HEPs) are indispensable for individual viability and development. However, experimental methods to identify HEPs are costly, time-consuming, and labor-intensive. Moreover, existing computational approaches often restrict predictions to the cell line level, failing to capture differences across human, cell line, and animal models.
Our Solution
We present PIC (Protein Importance Calculator), a sequence-based deep learning model fine-tuned on a pretrained protein language model. PIC substantially outperforms existing methods and provides comprehensive predictions at three complementary levels: human, cell line, and mouse.
History
-
Nov 2024PIC published in Nature Computational Science
-
Mar 2024PIC web server released
-
Nov 2023325 PIC models trained
-
Jun 2023PIC algorithm built
-
Feb 2023Original idea proposed
Contact us
Dr. Qinghua Cui
Dept. of Biomedical Informatics, Peking University Health Science Center
Email: cuiqinghua@bjmu.edu.cn
Citation
Kang, B., Fan, R., Cui, C., Cui Q. et al.
Comprehensive prediction and analysis of human protein essentiality based on a pretrained large language model.
Nature Computational Science, 2024.
Run Protein Prediction
Download Resources
General Prediction Results
Swiss-Prot essentiality scores for all human proteins.
all_general_prediction_results.csvSpecific Prediction Results
Cell line–level essentiality predictions across 323 human cell lines.
all_specific_prediction_results.csvCell Line Information
Metadata of 323 human cell lines, including tissue and disease origins.
all_cell_line_info.csvMicroprotein PES Scores
Predicted essentiality scores for microproteins.
all_microproteins_predicted_PES.csvPIC Source Code
Pretrained ESM-2 Models
Help
Introduction
Essential proteins are the expression products of essential genes that perform diverse biological functions closely associated with organismal growth and development. However, prevailing computational methods solely predict essential proteins from a single fixed level, thereby lacking a comprehensive approach to prediction and analysis. Here, we proposed Protein Importance Calculator (PIC), a sequence-based model designed for multi-level essential protein prediction. The overall workflow of our paper is shown below:

Visit PIC
PIC is available at http://www.cuilab.cn/pic.
Predict
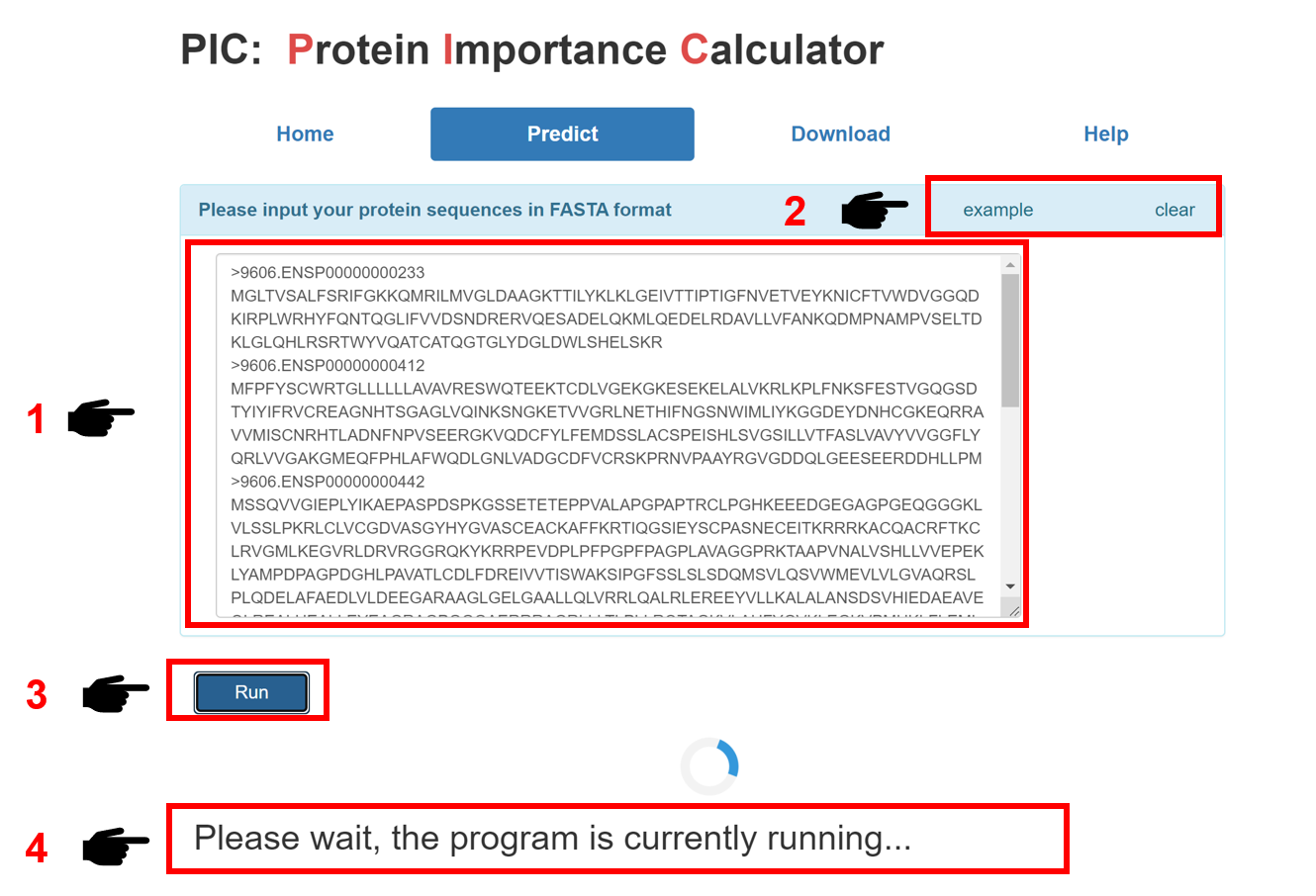
1. Input the interested protein sequences in FASTA format.
2. Click the 'example' button for example sequences.
3. Click 'Run' to submit your protein sequences.
4. Please be patient for a few minutes, and the prediction result page will appear soon.
Result
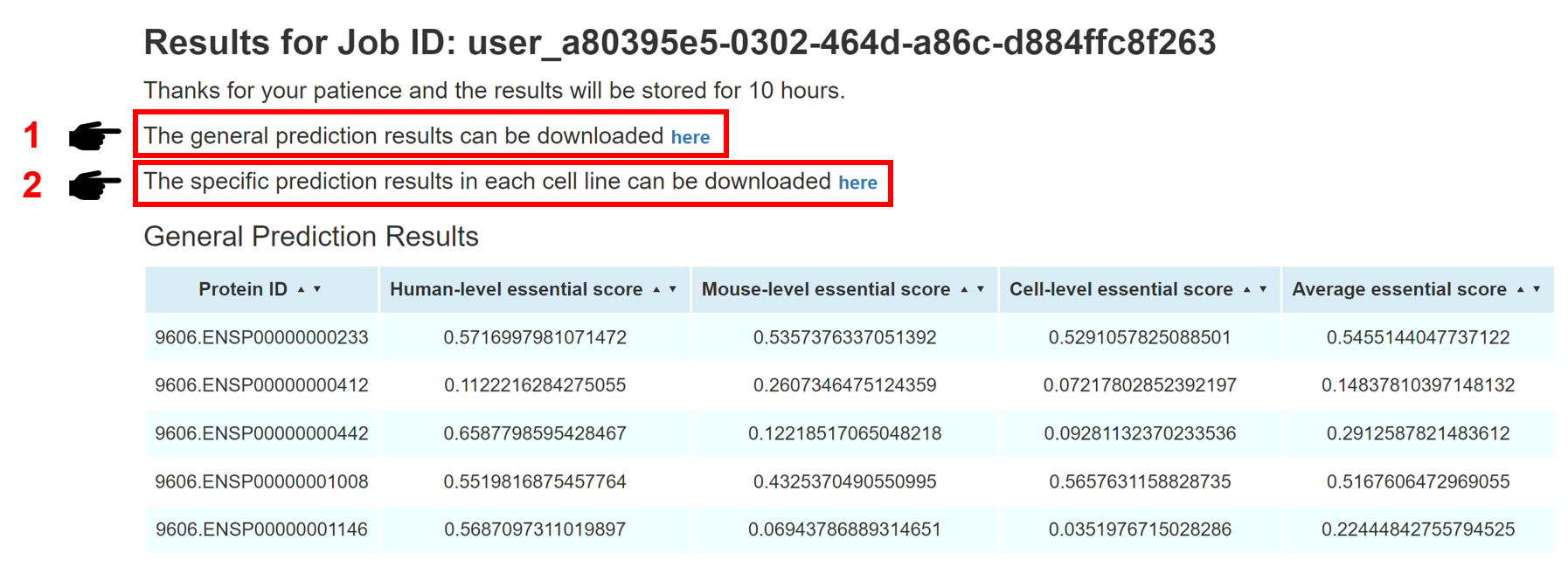
The prediction results will be shown in a few minutes. The general prediction results are presented in a table format, including Protein ID, Human-level score, Mouse-level score, Cell-level score, and Average score. Please refer to the Metric section for detailed meanings of each column. For the convenience of users, both general prediction results and specific prediction results can be downloaded in the result page. The general results file contains the overall prediction results from human-level, mouse-level, and cell-level. The specific results file provides predictions across 323 human cell lines.
Download
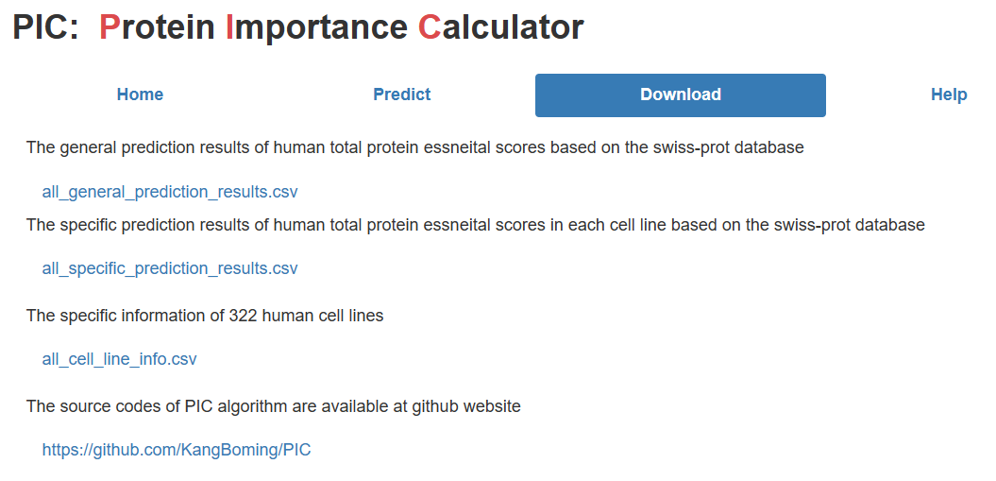
If you want to use the multi-level protein essential scores of all known human coding genes for large-scale data analysis, we provide the calculated essential scores of approximately 20,000 known proteins at multiple levels stored in CSV format files. Moreover, we also provide specific information of 322 human cell lines, including their tissue source and disease source. Finally, the PIC source code is available on GitHub, allowing anyone to access and extend the resource.
Metric Interpretation
In our paper, we anticipate that the essentiality of human proteins could be evaluated from three levels: (i) human-level: proteins rarely disturbed or truncated in the general population based on large-scale human genome sequencing. (ii) mouse-level: proteins whose knockout causes embryonic lethality in mice. (iii) cell-level: proteins whose knockout leads to decreased cell viability.
Therefore, a total of 324 basic PIC models were trained: one for human-level (PIC-human), one for mouse-level (PIC-mouse), and 323 for cell-level. Each cell-level model was treated as a basic classifier, and a soft voting strategy was employed, aggregating the mean probability values across 323 cell lines. This process resulted in a high-performance classifier represented by PIC-cell.
We defined the probability values output by PIC-human, PIC-mouse, and PIC-cell models as the essential scores at their respective levels. To create a comprehensive metric, we averaged the three-level essential scores as the average essential score, thus providing an integrated evaluation of protein essentiality across human, mouse, and cell perspectives.