HMDD v4.0 Tutorial
June, 2023
MicroRNAs (miRNAs) are one class of important regulatory noncoding RNAs, which is ~22nt in length and mainly repress gene expression at post-transcriptional level. Emerging evidence has shown that miRNAs play critical roles in various important biological processes and therefore the dysfunctions related with miRNAs are often associated with human disease, including cancer and cardiovascular disease. Therefore, miRNAs are representing one class of important biological molecules in understanding the mechanisms of disease formation and development, and in identifying biomarkers for disease diagnosis and treatment.
HMDD (the Human microRNA Disease Database) is a resource that curates experimentally supported miRNA and disease association data. HMDD was originally constructed on December 2007 and was updated more than 40 times during the past fifteen years. On June 2013, we released HMDD version 2 (HMDD v2.0). In the version 2, we curated miRNA-disease association data in more details, such as data from genetics, and epigenetics, data from samples of circulation (such as blood, serum, and plasma etc), and data from miRNA-target interactions. On June 2018, we released HMDD v3.0. Now in the latest version 4, we have classified the literature evidence in more details, which results in 8 evidence classes in the following figure. Currently, users can browse and search the database by the names of miRNAs and diseases. Moreover, users can download all data in HMDD v4.0 (The whole dataset of miRNA-disease association data in the 'Download' page).
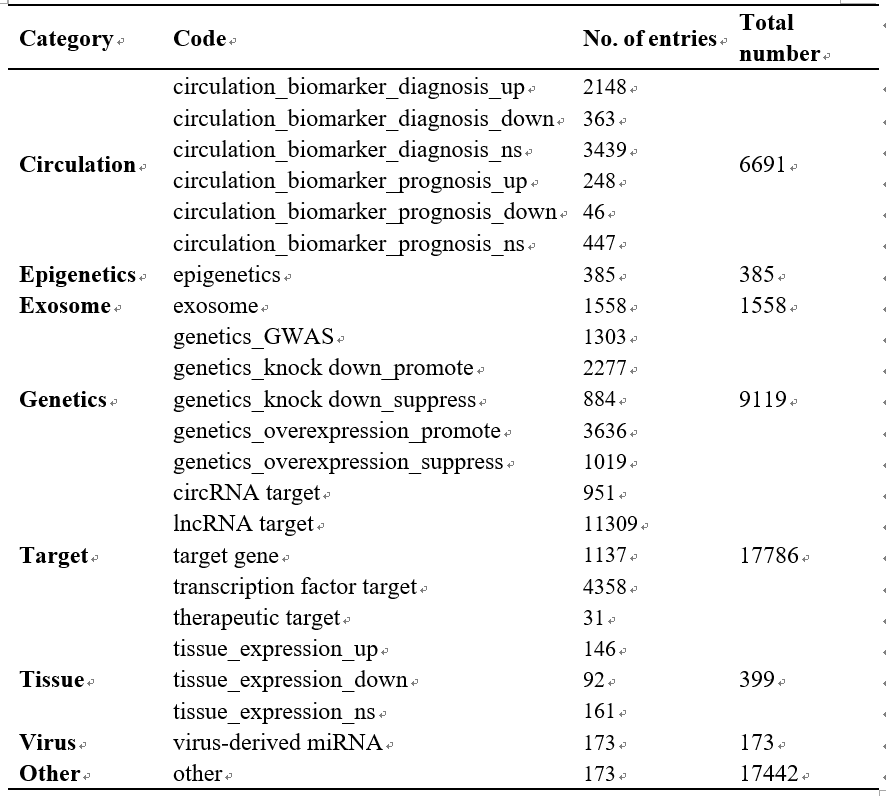
Note: Two of these categories, 'Exosome' and 'Virus', are newly introduced. 'Exosome' is collected according to whether the miRNAs are derived from exosome involved in human diseases, and ‘Virus’ represents the virus-derived miRNAs associated with human diseases.
The detailed usage of the database is as followings:
[1] Visiting HMDD v4.0: Log on HMDD at http://www.cuilab.cn/hmdd
[2] To browse miRNA-disease association data in the database, please click the menu "Browse" (1 in Figure 1). For example for the miRNA-disease association data from miRNA-target interactions, select “Target” (step 1). To browse the entries related to the candidate disease, please click “disease” (step 2) and select the one you are interested in (step 3). To browse the entries related to the candidate miRNA, please click “miRNA” and select the one you are interested in. For other types of miRNA-disease association data, please select specific browse options, such as “genetics”, “epigenetics”, and “circulation”.In addition, we added the third party annotations about miRNA and disease; users can click the miRNA/disease name to see these annotations (step 4). The result will be shown in the right panel. Clicking “click to download” to download the browsing result (step 5).
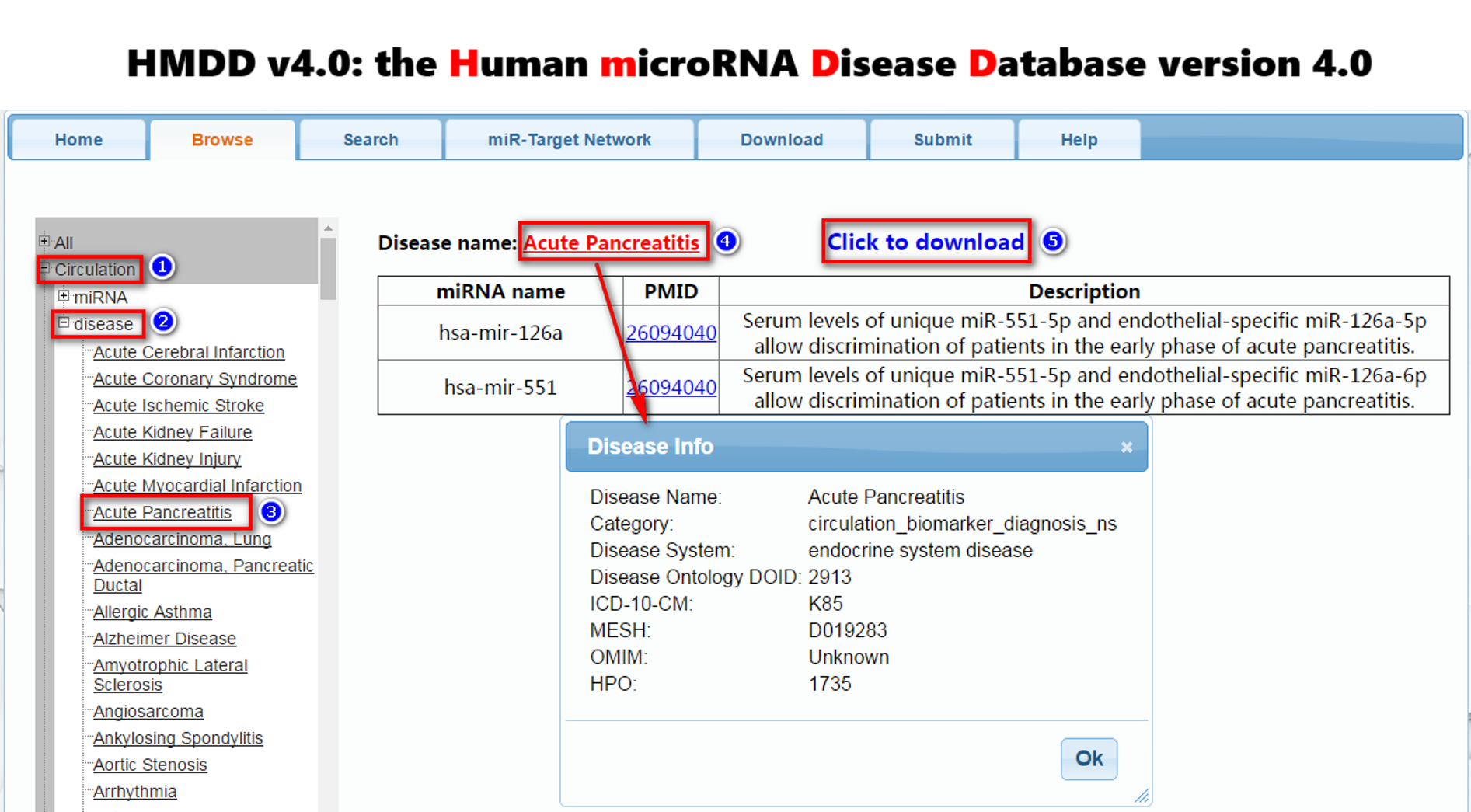
Figure 1. A demo for browsing data in the database
[3] To search data in the database, select the menu "Search". HMDD v4.0 provides functions of "search" by full or partial names of miRNAs or diseases. Input your candidate keywords into corresponding blanks and submit the query.
Each “search” will return entries including the following four items.
(1) miRNA name
(2) Disease name
(3) Reference –The PMID links to the reference in PubMed.
(4) Description - detailed description of the miRNA-disease association.
[4] To view the networks between the disease-associated miRNAs and disease-associated genes, select the menu “miR-Target Network”. If the user clicks one miRNA name, the disease-associated genes it targets will be displayed. If the user clicks one disease name, the network between miRNAs and genes associated with this disease will be shown.
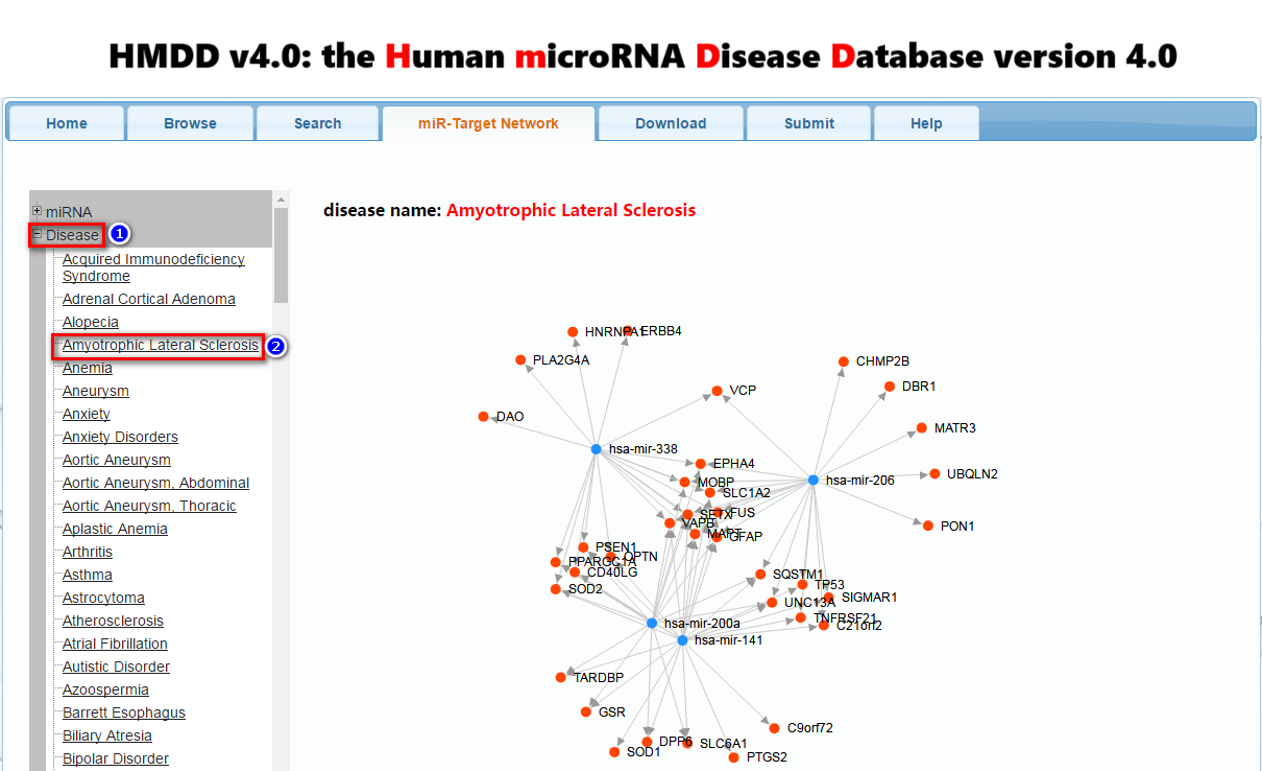
Figure 2. A demo for viewing the networks between the disease-associated miRNAs and disease-associated genes
[5] To download data in the database, select the menu "Download". HMDD v4.0 provides two formats of downloadable file in TEXT and Excel formats, respectively. The users can download all data in HMDD v3.0, including the experimentally supported miRNA-disease association data, miRNA-disease association data from genetics, miRNA-disease association data from epigenetics, miRNA-disease association data from circulation samples, and miRNA-disease association data from miRNA-target interactions.
[6] To submit new entries to the database, select the menu "Submit". The users need to input their data into corresponding blanks and then submit the query. We will further curate the submitted information to determine whether to add the new entries to the database or not.
[7] Currently, all data in HMDD is free only for academic users and please contact Dr. Qinghua Cui for commercial using. If you used the data in HMDD v4.0, please cite "Lu M, Zhang Q, Deng M, Miao J, Guo Y, et al. (2008) An Analysis of Human MicroRNA and Disease Associations. PLoS ONE 3(10): e3420.", "Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014 Jan;42(Database issue):D1070-4." and "Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, Zhou Y, Cui Q. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019 Jan 8;47(D1):D1013-D1017."
If there are any comments or suggestions, please contact Dr. Qinghua Cui at cuiqinghua@bjmu.edu.cn or cuiqinghua@hsc.pku.edu.cn.



